Small angle X-ray scattering (SAXS) reveals solution structures of biological macromolecules and synthetic nanoparticles at 1-2 nm resolution.
SAXS is a universal technique applicable to a broad range of particle sizes, from small peptides to huge macromolecular machines with molecular weight from about 5 kDa up to 100 MDa. With the brilliant synchrotron sources, robotic sample changers and novel approaches to reconstruct 3D models, SAXS became a major tool to rapidly and comprehensively characterize macromolecular and nanostructured systems.
The main principles of SAXS were developed in the late 1930s by A. Guinier with his studies of metallic alloys. Already in the first monograph on SAXS by Guinier and Fournet (1955) it was demonstrated that the method yields not just information on the sizes and shapes of particles but also on the internal structure of disordered and partially ordered systems.
Conceptually, a SAXS experiment is simple: a sample is illuminated by X-rays and the scatteried radiation is registered by a detector. As the SAXS measurements are done very close to the primary beam ("small angles"), the technique profits immensely from the brilliance of X-ray photon beams provided by particle accelerators known as synchrotrons. Europe has nearly 20 of these huge machines, including new "third-generation" or "high brilliance" designs that produce especially intense, narrow X-ray beams.
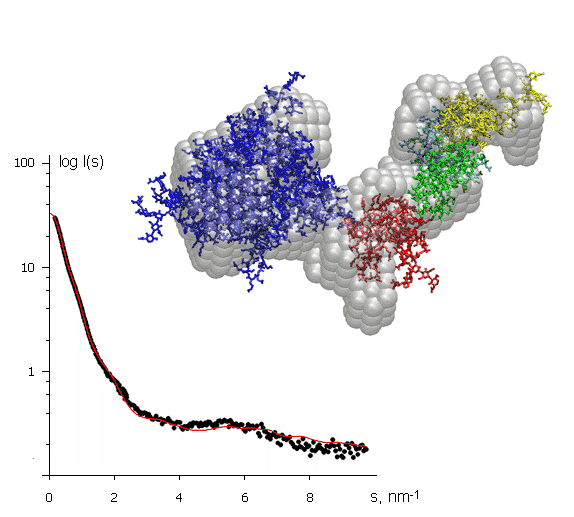
The X-ray scattering curve on the left (intensity versus scattering angle) was used to create a low-resolution model of a protein, shown in grey. Superimposed on this (in colour) are atomic models of separate domains obtained from crystallography and positioned to fit the SAXS data. The protein molecule is about 10 nm across. This example illustrates how SAXS can help to assemble together high resolution models of individual domains provided by protein crystallography into the model of the entire macromolecular complex.